Principal Investigator
Mark WronkiewiczEmail: wronk@jpl.nasa.gov
Project Overview
The Ocean Worlds Life Surveyor (OWLS) project is focused on searching for water-based life in our solar system. Ocean worlds are a natural target for scientific inquiry since all life we know of depends on liquid water. The OWLS team is developing several instruments that can detect signs of microorganisms (think single cell bacteria or eukaryotes) in liquid water samples. These will include microscopes that record videos (so we can literally look for swimming cells) as well as a mass spectrometer (to search for chemical building blocks that support life).
Europa and Enceladus – moons of Jupiter and Saturn, respectively – are two ocean worlds that the scientific community is interested in. Unfortunately, both are hundreds of millions of miles away. Because of their distance to Earth, transmitting data from those locations is very challenging and requires a lot of electricity. Our total data transmission budget is so limited that we’ll only be able to transmit ~0.01% of the raw data we collect back to Earth! To overcome this obstacle, MLIA is building autonomous software that can analyze, summarize, and prioritize data from these instruments so we can select the best data snippets to transmit back for scientific analysis. Our autonomous software will eventually enable science at a distance that wasn’t possible just a few years ago.
The OWLS autonomy team is leveraging the Labelbox platform to capture machine learning labels. Check out their recent blog post on this collaboration here.
Autonomy for Individual Instruments
Mass Spectrometer
NASA has a long history of searching for the chemical building blocks that underlie biology. The OWLS project will follow in these footsteps by using a mass spectrometer to assess the chemical compounds present in our water samples. Mass spectrometry measures the masses of particles and molecules within a sample – in our case, a small amount of water. These measurements serve as a fingerprint to help scientists determine the particular molecules present. We will particularly be searching for molecules like amino acids, peptides, nucleic acids because they support all biological organisms we know about.
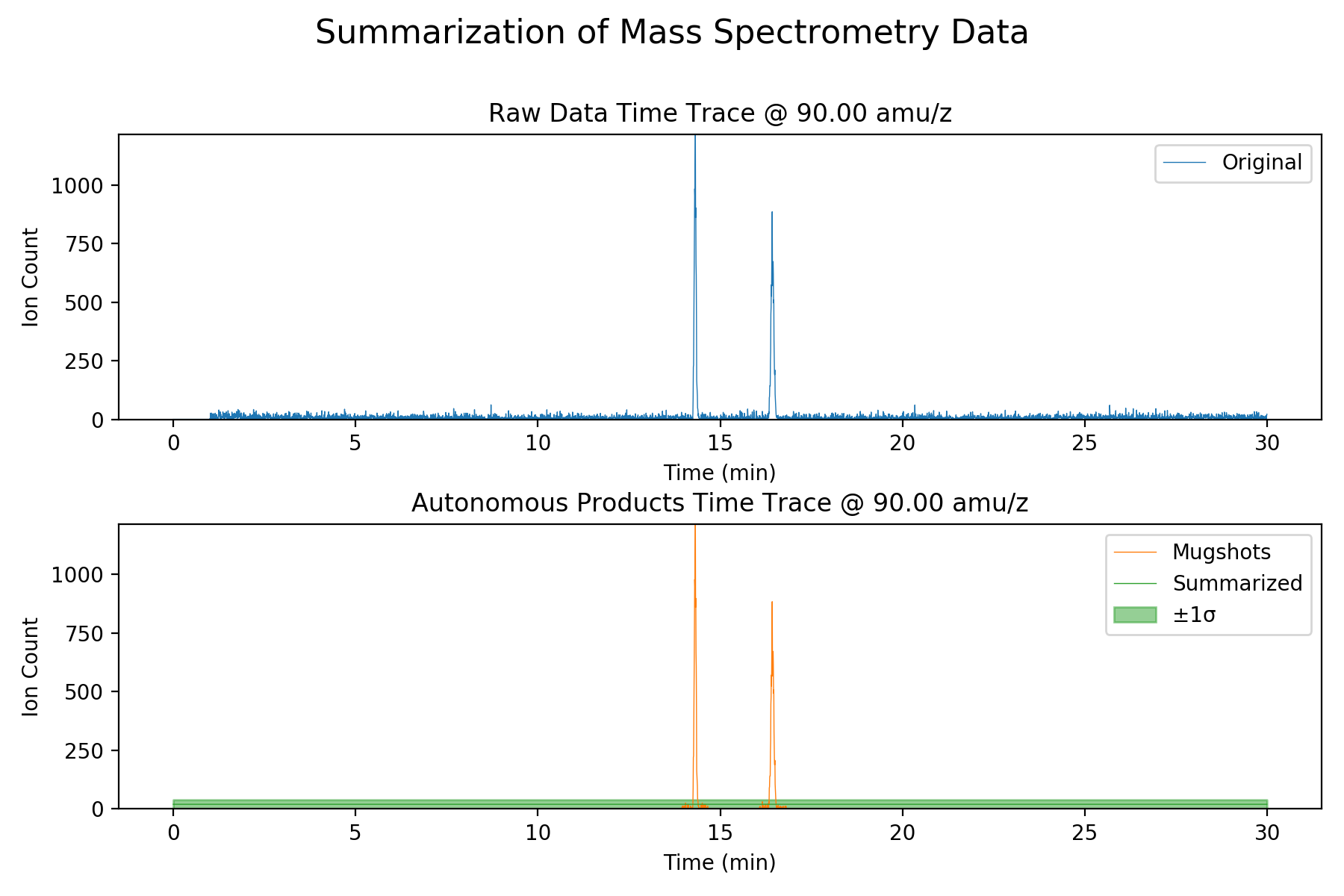
Top: A slice of mass spectrometry data. The position of the peaks and their heights can be interpreted to determine which compounds are present in the sample.
Bottom: A slice of data reconstructed from an autonomous summary. The raw data is too large to downlink in its entirety, so the system detects and downlinks only the data around the peaks. The rest is summarized as a mean and standard deviation to provide context.
Holographic Microscopy
Besides searching for chemical signatures of life, we will search directly for swimming microorganisms in microscopy videos. The OWLS project will investigate water samples using several different microscopes including a Digital Holographic Microscope (DHM). This instrument records video over tens of seconds and provides us with a 3D representation of the sample volume rather than the conventional 2D light-field microscope typically used in a biology class. Our software tracks and analyzes the movement patterns of particles within these videos. We then use a machine learning model to separate life-like particles moving under their own power from background debris that’s floating passively through the microscope field. Once we’ve identified microorganisms, we compile snapshots and other data that would be returned to Earth.
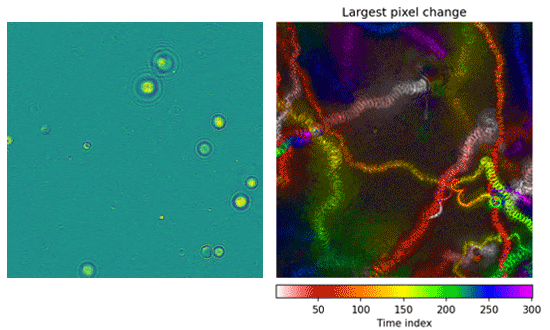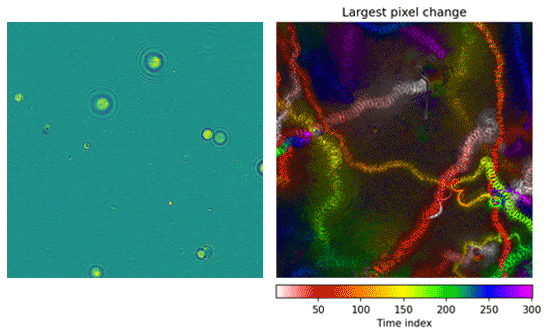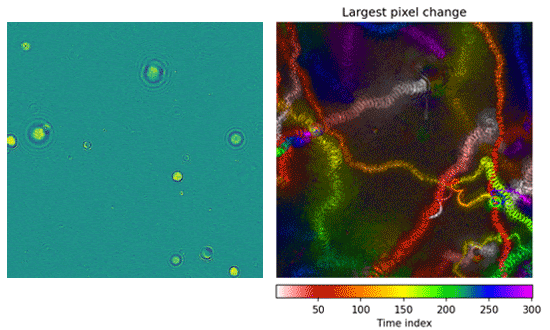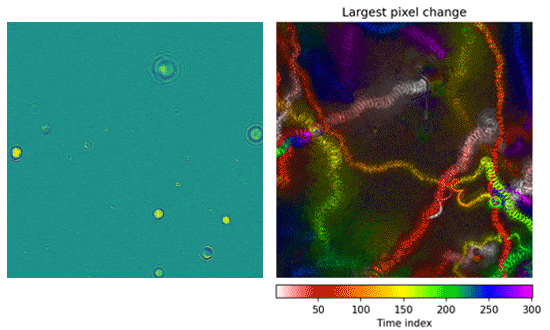
Left: Microorganisms swim around within the microscope’s field of view. Right: A motion history image illustrates where in time motion occurred. The presence of streaks and paths is a simple, human-interpretable way to see that this experiment contained swimming microorganisms.
Fluorescence Imaging
Along with the mass spectrometer, we have another microscope that can also search for the chemical building blocks of life. Certain large molecules like lipids and nucleic acids are associated with every living thing we know. We will search for some of these macromolecules using the Volume Fluorescence Imager (VFI), which relies on dyes to bind to these macromolecules. The dyes themselves are fluorescent so they will appear to “glow” within the VFI once excited by a laser. If we introduce these dyes to the water sample and see bright regions or globs within the VFI, this indicates that there’s a concentration of these potentially life-supporting macromolecules. The DHM and VFI will actually share the same field of view meaning we can look for overlap of life-like movement and life-supporting macromolecules in the same sample at the same time.
Data Prioritization
Science instruments for planetary missions are becoming increasingly advanced and able to collect ever larger volumes of science data. Limited downlink bandwidth, especially for missions to the outer planets, means that data must be carefully selected for return to Earth in order to maximize mission productivity and science yield. The Joint Examination for Water-based Extant Life (JEWEL) system is designed for the Ocean Worlds Life Surveyor (OWLS) instrument, which will use a combination of microscopy and mass spectrometry to search for signs of life on icy worlds such as Europa and Enceladus. Because signs of life are likely rare relative to the large quantities of raw data these instruments will collect, we have developed systems that will analyze data onboard the spacecraft to produce smaller summary products called autonomous science data products (ASDPs). These ASDPs include the tracks of moving particles in raw microscope images and the peak locations and sizes within mass spectra.
Given a set of candidate ASDPs available for downlink, the goal of this work is to prioritize these products to determine the downlink order while balancing two objectives: (1) maximize the estimated science utility of the downlinked products and (2) maximize the diversity of the products downlinked to avoid redundancy and enable the discovery of rare science content.
